Before you get into dealing with the minutia of database performance, you have to stand back and ask the obvious questions like: “Do I actually need all this data?”

Even with the cleverest querying or aggregation, fine tuning indexes and studying execution plans, you are unlikely to make the same sort of gains that you can from reducing your working collection to just include the information you need.
Also, much of the black art of OLAP querying is that of eliminating the need to repeatedly make the same joins for all your queries when the underlying data isn’t constantly changing.
To effectively reduce your working data so as to optimize its performance, you must first understand the nature of the data.
If it is changing the whole time, for example, then it requires rather different treatment to the way that you’d tackle static data.
Few developers will want to admit that a lot of their data is unchanging, but the whole accounting process from ancient times is based on the idea that past transactions don’t change in any way.
In fact, a lot of our data can be summarized or re-aggregated only if a correction is made to the data that alters a total.
Why then search through data about past events on every report to get information that you already know?
The trick is to archive what you don’t want, and just extract and aggregate the summary of the data that you need.
To do this, you need to get to grips with an aggregation or grouping process that will, in effect, group the historic data into the smallest slices that you’re ever likely to need, and for the odd unusual query, you still have the actual data of the individual documents or records that you can return to.
To illustrate this, we need a database to try experiments on. I find more pleasure in manipulating real data than the usual fake data, so, to demonstrate the basics, we will use as a demonstration a taxonomy of marine mammals, some of the most interesting and engaging of creatures.

Marine mammals, feat. Studio 3T’s very own Hugh the Manatee
We were very privileged to be given part of the WORMS (World Register of Marine Species) database to do this. It changes very little, so we are safe in being lavish in extracting just what we want.
Querying the Marine Mammals database
Our objective is to get a list of marine mammals, with their English names where possible, indented to show their taxonomy.
Normally, I’d be feeding this into a tool for tracing relationships and taxonomy or for plotting. However, this simpler list is a neat objective for our purposes as we can do everything in the MongoDB GUI, Studio 3T.
A document from the WORMS database will provide 23 fields, but for our purposes we will only need a subset of this.
A typical document looks like this:
{
"_id" : ObjectId("5a5df4ec3185824860a6a6db"),
"taxonID" : NumberInt(383653),
"scientificName" : "Hunterus temminckii",
"scientificNameAuthorship" : "Gray, 1864",
"acceptedNameUsageID" : NumberInt(220222),
"acceptedNameUsage" : "Eubalaena australis",
"taxonRank" : "Species",
"kingdom" : "Animalia",
"phylum" : "Chordata",
"class" : "Mammalia",
"order" : "Cetartiodactyla",
"family" : "Balaenidae",
"genus" : "Hunterus",
"specificEpithet" : "temminckii",
"taxonomicStatus" : "unaccepted",
"taxonRemarks" : "synonym",
"modified" : "2009-03-05",
"scientificNameID" : "urn:lsid:marinespecies.org:taxname:383653",
"parentNameUsageID" : NumberInt(383388),
"bibliographicCitation" : "Perrin, W. (2009). Hunterus temminckii Gray, 1864. In: Perrin, W.F. (2017).
World Cetacea Database. Accessed through:
World Register of Marine Species at http://www.marinespecies.org/aphia.php?p=taxdetails&id=383653",
"isMarine" : true,
"isFreshwater" : false,
"isTerrestial" : false
}
Before doing any serious querying we need to get a feel for the database and how it is arranged.
By using simple method (the distinct() method ) on the collection, we can explore categories such as the taxonomic rank (“taxonRank”).
db.marineMammalSpecies.distinct( "taxonRank" )
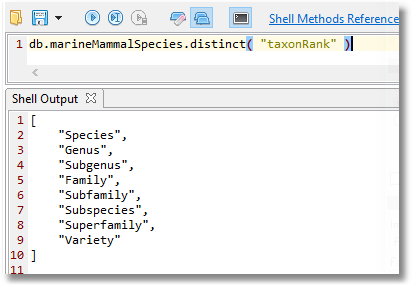
This tells us that we have a collection of documents, each one of which describes, according to the taxonomic rank (“taxonRank”) either the Family, Superfamily. Subfamily, Species, Genus, Variety, Subgenus or Subspecies of a marine mammal.
However, this isn’t really enough information. This doesn’t tell you how many documents of each taxonomic rank that there are. This MongoDB aggregation will provide this information.
db.marineMammalSpecies.aggregate([
{
$match: { //only include documents with a taxonrank
taxonRank: { $not: {$size: 0} }
}
},
{
$group: { //count how many of each type (using Lowercase name)
_id: {$toLower: '$taxonRank'},
count: { $sum: 1 }
}
},
{ $sort : { count : 1} } //sort from smallest to biggest count
]);
This tells you what all these documents represent. We have the names for all parts of the taxonomy here so we can plot the entire taxonomy just from the data.
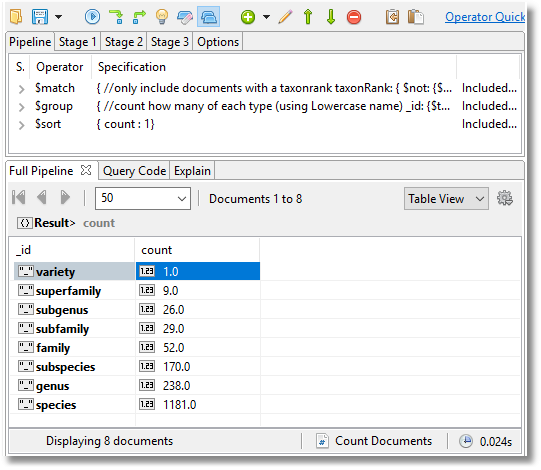
Not all of these documents describe the current name of the species since the taxonomy evolves with our understanding of the evolution of these sea creatures. We have all the previous names and the names that are no longer in use.
We have work to do before we can do our final querying because we are likely to be concentrating on the current taxonomy, and we aren’t interested yet in the more esoteric data such as the bibliographic citation for a name.
We therefore need to reduce the data to what we need and create a new collection.
We don’t want the names that are no longer current, but we want the zoological names to be accompanied by the vernacular name such as Seal, Whale, Porpoise or Manatee.
Creating the MongoDB aggregation that produces the working collection
First, we see how to filter just the documents that refer to the current nomenclature.
An inspection of the data suggests to us that if a collection has a “acceptedNameusageid” attribute, and if this is the same as the ‘taxonID”, then it is likely to be a currently accepted name for part of the taxonomy.
We can produce a stage of an aggregation query to determine this:
Stage 1
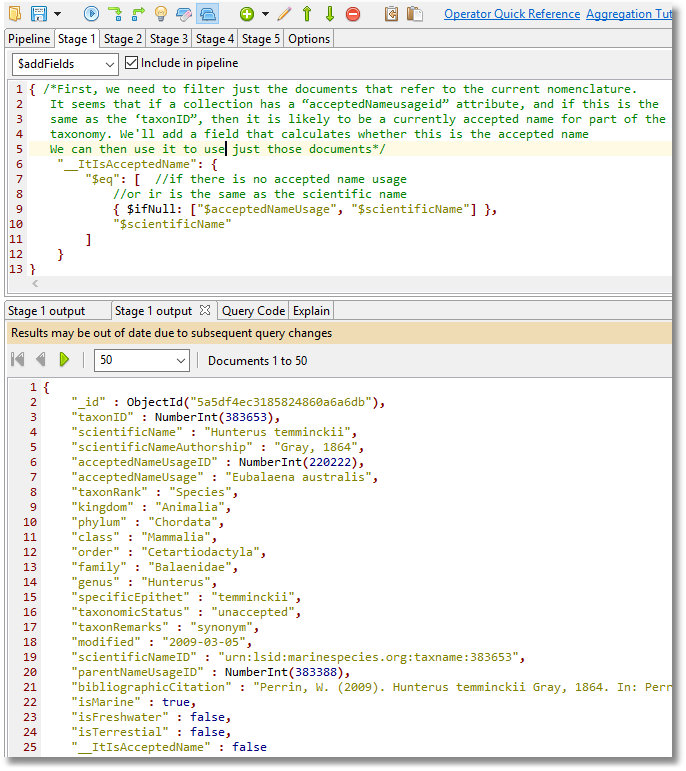
When we try this out we find that the output of the stage gives us a collection that are seemingly valid but have a taxonomicStatus of ‘nomen dubium’.
Could we use this for filtering? If so, what values? We can see what range of values is in the taxonomicStatus in the collection by means of …
db.marineMammalSpecies.distinct( "taxonomicStatus" ) [ "unaccepted", "accepted", "alternate representation", "nomen nudum", "nomen dubium", "temporary name" ]
We can eliminate very easily the ones that aren’t accepted if we want to by adding a condition to the next match phase.
Stage 2
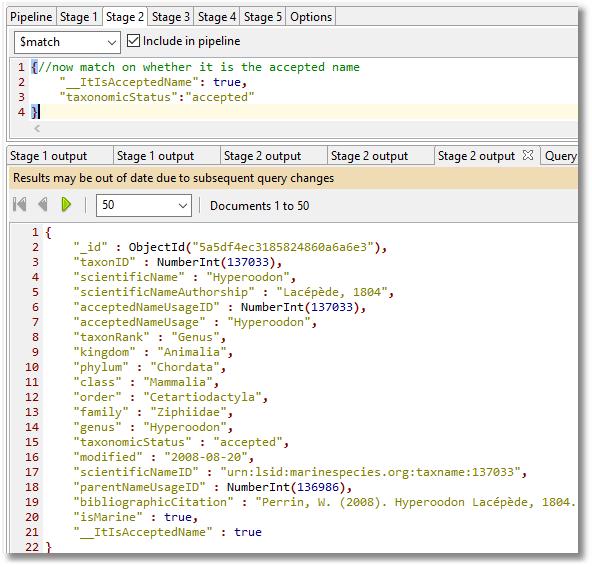
This is a start, but it would be good to attach vernacular names to these, so we get an idea of whether we are dealing with dolphins, porpoises, whales, walruses or whatever.
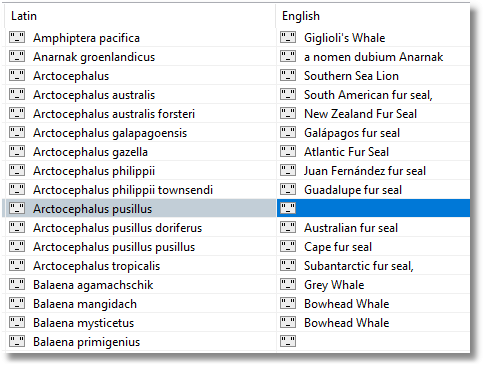
We next nip off to the Wikipedia and any other sources we can find in order to get hold of a list of the common names of these mammals and their Latin specific epithets. We need to have this for the name of the order, the Family, the Genus and the Specific Epithet.
It isn’t a good idea to add fields to someone else’s collection because this makes updates more complicated when they update it. We need to create a separate collection in order to store our vernacular names.
We can then join to this collection in order to create a new collection that is easy to query and holds all the information we will require.
{
"_id" : ObjectId("5a86f267318582208819a269"),
"Latin" : "Arctocephalus australis australis",
"English" : "South American fur seal "
},
{
"_id" : ObjectId("5a86f267318582208819a348"),
"Latin" : "Arctocephalus pusillus pusillus",
"English" : "Cape fur seal"
},
{
"_id" : ObjectId("5a86f267318582208819a349"),
"Latin" : "Arctocephalus pusillus doriferus",
"English" : "Australian fur seal"
},
In Studio 3T this collection looks like this, and we can edit the individual entries:
Stage 3
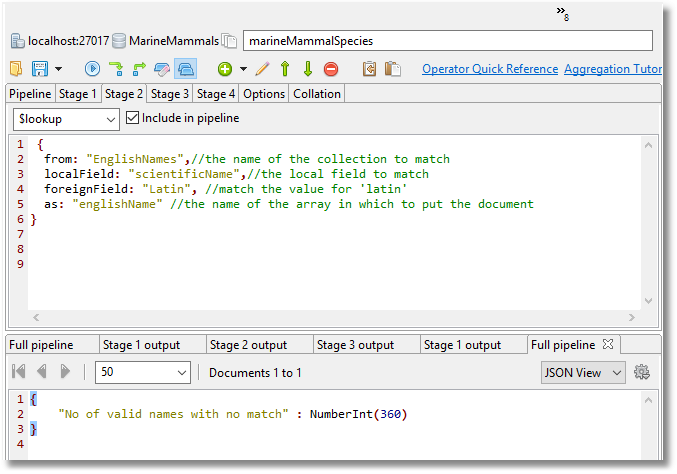
The next stage in the MongoDB aggregation that we are preparing is to add the English names with a simple left outer join.
MongoDB allows you to do left outer joins via the lookup() stage which joins to another collection in the same database to filter in documents from the “joined” collection for processing.
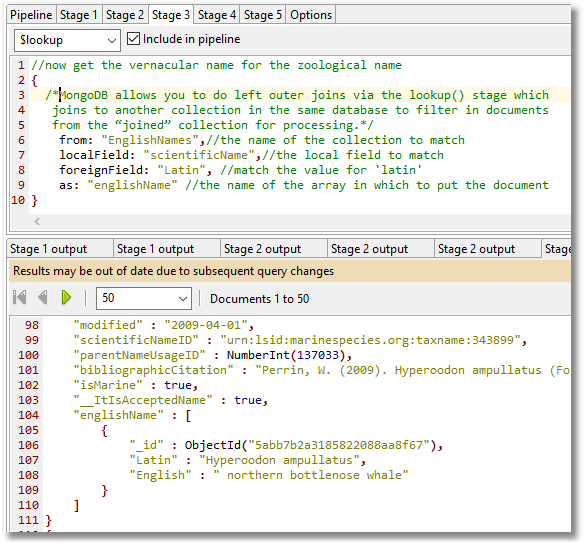
You can also do right outer joins, and inner joins, because they can be derived from the left outer join.
The inner join is just a left outer join where there was a successful match, and so if we test for a successful join, this provides you an inner join, telling you all the species and subspecies that have a match.
By doing the check that the English name exists in the left outer join, it becomes an inner join.
db.marineMammalSpecies.aggregate([
{
$lookup: //get the vernacular name for the zoological name
{
from: "EnglishNames",//the name of the collection to match
localField: "scientificName",//the local field to match
foreignField: "Latin", //match the value for 'latin'
as: "englishName" //the name of the array in which to put the document
}
}, {
"$match": {
"englishName.English": { "$exists": true }
}
}
])
Stage 4
Now we’ve got all of this, it is time to reduce the WORMS database to include just the items we want and combine this with the English name which the previous stage left as a new array field from the lookup operation whose elements are the matching documents from the “joined” vernacular (English) name collection.
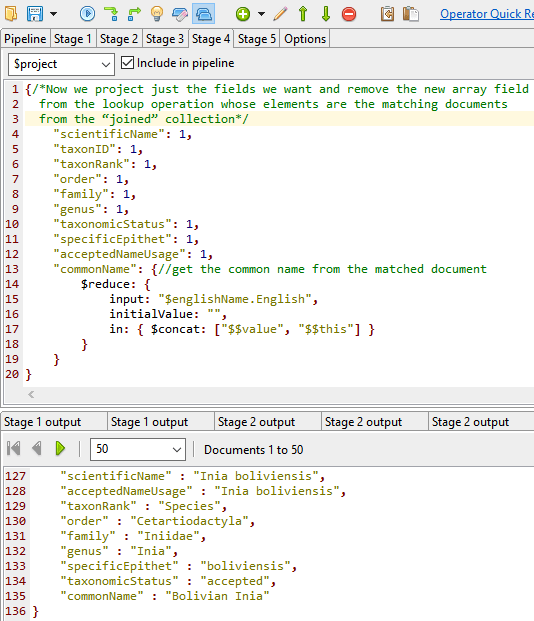
The full MongoDB aggregate query
We now have an aggregate query that we can use to create a new collection called validSpecies with just the current names of the species and subspecies.
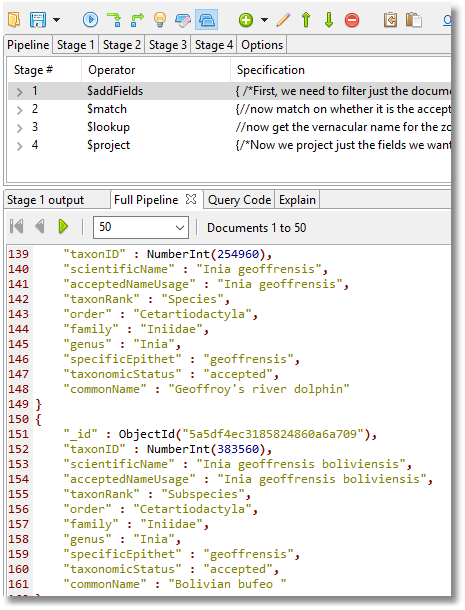
db.validSpecies.insert(db.marineMammalSpecies.aggregate(
// Pipeline
[
// Stage 1
{
$addFields: { /*First, we need to filter just the documents that refer to the current nomenclature.
It seems that if a collection has a acceptedNameusageid” attribute, and if this is the same as the
‘taxonID”, then it is likely to be a currently accepted name for part of the taxonomy. We'll add a
field that calculates whether this is the accepted name We can then use it to use just those
documents*/
"__ItIsAcceptedName": {
"$eq": [ //if there is no accepted name usage
//or ir is the same as the scientific name
{ $ifNull: ["$acceptedNameUsage", "$scientificName"] },
"$scientificName"
]
}
}
},
// Stage 2
{
$match: {//now match on whether it is the accepted name
"__ItIsAcceptedName": true, "taxonomicStatus":"accepted"
}
},
// Stage 3
{
$lookup: //now get the vernacular name for the zoological name
{
/*MongoDB allows you to do left outer joins via the lookup() stage which joins to
another collection in the same database to filter in documents from the “joined”
collection for processing.*/
from: "EnglishNames",//the name of the collection to match
localField: "scientificName",//the local field to match
foreignField: "Latin", //match the value for 'latin'
as: "englishName" //the name of the array in which to put the document
}
},
// Stage 4
{
$project: {/*Now we project just the fields we want and remove the new array field
from the lookup operation whose elements are the matching documents from the “joined” collection */
"scientificName": 1, "taxonID": 1, "taxonRank": 1, "order": 1,"family": 1,
"genus": 1, "taxonomicStatus": 1, "specificEpithet": 1, "acceptedNameUsage": 1,
"commonName": {//get the common name from the matched document
$reduce: {
input: "$englishName.English",
initialValue: "",
in: { $concat: ["$value", "$this"] }
}
}
}
},
]
).toArray());//put in a new collection
Doing joins to check our work
Now we can check which English names are missing in case we are keen to improve the collection (in fact, some species just don’t have real English names).
This requires a left outer join with a NULL check, in relational terms. we match only the unsuccessful documents. This is much the same as the logic is SQL.
db.validSpecies.aggregate([
{
"$project": {
"scientificName": 1,
"taxonomicStatus": 1,
"_id": 0
}
}, {
$lookup: //get the vernacular name for the zoological name
{
from: "EnglishNames",//the name of the collection to match
localField: "scientificName",//the local field to match
foreignField: "Latin", //match the value for 'latin'
as: "englishName" //the name of the array in which to put the document
}
}, {
"$match": {
"englishName.English": { "$exists": false },
"taxonomicStatus": "accepted"
}
}
])
How many English names are missing? This is the same join but with the equivalent of the SQL count(*) aggregation. We can produce a more efficient aggregation by first filtering for just the fields we need:
db.validSpecies.aggregate([
// Pipeline
[
// just aggregate the fields we need
{
$project: {//only include the fields you need to make this index-friendly
"scientificName": 1, //we need to join to this
"taxonomicStatus": 1 //we need to test this
}
},
// Stage 2
{
$lookup: {
from: "EnglishNames",//the name of the collection to match
localField: "scientificName",//the local field to match
foreignField: "Latin", //match the value for 'latin'
as: "englishName" //the name of the array in which to put the document
}
},
// Stage 3
{
$match: { //only include those whose lookup was successful
"englishName.English": { "$exists": true }
}
},
// Stage 4
{
$count: "No of valid names with no match"
},
]
// Created with Studio 3T, the IDE for MongoDB - https://studio3t.com/
);
So with some help from the good people behind the WORMS database and Wikipedia to find the English names, and these queries I’ve shown you to check progress, it looks as if we can now assemble a fairly representative list.
Showing the taxonomy
Now we have a new, cleaner, filtered collection with just the information we want, and no need to complicate further queries with $lookup or $reduce.
Depending on your MongoDB database, there will be a lot of cleaning and adjusting you can do to make life simpler for later ad-hoc queries.
db.Taxonomy.insert(db.validSpecies.aggregate([
{
$project://Reshapes each document in the stream, to provide a uniform document.
{
theOrder: { $ifNull: ["$order", " "] }, // for sorting
theFamily: { $ifNull: ["$family", " "] },// for sorting
theGenus: { $ifNull: ["$genus", " "] },// for sorting
theSpecificEpithet: { $ifNull: ["$specificEpithet", " "] },// for sorting
"taxonRank": 1, //means include this
"taxonomicStatus": 1, //means include this
"_id": 0, //do not include this
"Line": { //This field is provided to produce the indentation
$concat: [ //provide indentation according to its order in the taxonomy
{
$switch:
{
branches: [
{
case: { $eq: ["$taxonRank", "Superfamily"] }, then: ""
}, {
case: { $eq: ["$taxonRank", "Family"] }, then: " "
}, {
case: { $eq: ["$taxonRank", "Subfamily"] }, then: " "
}, {
case: { $eq: ["$taxonRank", "Genus"] }, then: " "
}, {
case: { $eq: ["$taxonRank", "Subgenus"] }, then: " "
}, {
case: { $eq: ["$taxonRank", "Species"] }, then: " "
}, {
case: { $eq: ["$taxonRank", "Subspecies"] }, then: " "
}
],
default: "$taxonRank" //for debugging. This shouldn't happen!
} // and display the common name
}, "$scientificName", { $cond: { if: { $eq: ["$commonName", ""] },
then: "",
else: { $concat: [" (", "$commonName", ") "] } }
}
}
}
}, {
$match: { //only include those whose name is accepted.
taxonomicStatus: "accepted"
}
} // sort the list according to their location in the taxonomy
{ $sort: { theOrder: 1, theFamily: 1, theGenus: 1, theSpecificEpithet: 1 } },
// and only project the indentation, the scientific name and the common name
{ $project{ theOrder: 0, theFamily: 0, theGenus: 0, theSpecificEpithet: 0, taxonRank: 0,
taxonomicStatus: 0 } }
]).toArray()
Now, all we need to do to see the taxonomy is to execute this:
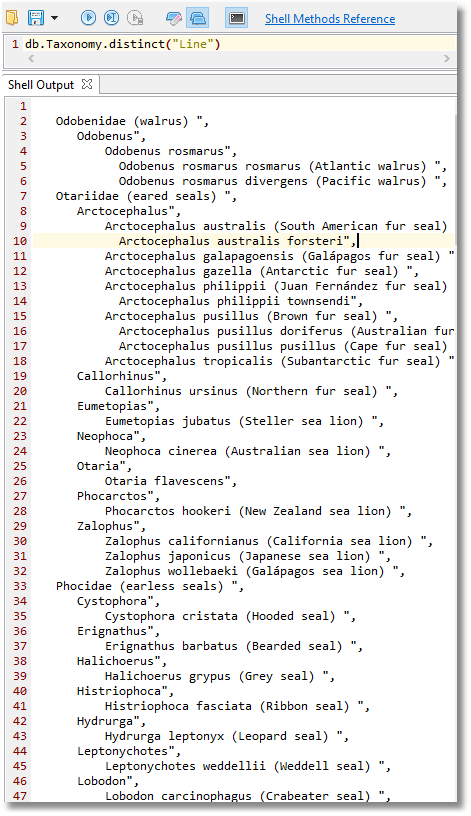
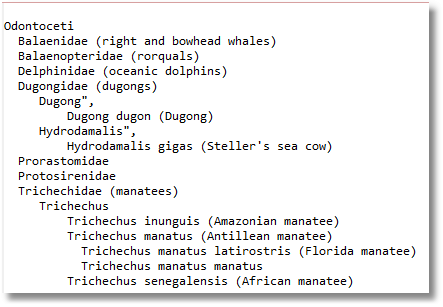
..but let us not forget the noble manatee towards the end of the result:

If I were going on to process this information in reality, I would be using a package that is designed to represent taxonomies, so the way of rendering the results would depend on what was required for input.
GraphViz is an old but familiar way of doing this. This will allow you to plot each type of name (such as Family, genus and species) in a different way, and all you need to do is to specify the relationships in Graphviz dot language. Hmm. Maybe that is a good exercise!
Conclusions
One of the best ways of increasing the performance of a MongoDB database is to stand back and look at the data you are using. Is all your data constantly changing? Why are you scanning through all the data otherwise?
I’ve come across production database systems that constantly ran queries on previous years’ data even though it was obvious that it would never, and should never, change.
Aggregation collections are like explicit indexes because they allow reporting to take place without having to scan the original data.
If you then go on to index these aggregations your queries will fly, and as the data is generally highly reduced in size, you can be lavish with indexes to cover every common query. Fresh data can then be added in without affecting query performance much.
These MongoDB aggregations take care to prepare, and they take some experimentation to get right; however, they could be the most effective way of increasing MongoDB database performance.

Thank you to the folks at WoRMS (World Register of Marine Species), whose database we queried to illustrate the power of MongoDB aggregation.